Binary Upper Triangular Matrix (BUTLR) File Format

The Binary Upper TrianguLar MatRix, or BUTLR (.butlr, .btr), file format encodes contact matrices derived from chromosome conformation capture (3C)-based technology (i.e. Hi-C). The BUTLR format adds binary indexed encoding, allowing the random access of contact matrices and enhancing performances in both computing time and memory. In the 3D Genome Browser, such random access is especially important, as it enables the remote access of contact matrix files hosted on other servers, directly querying the region of interest instead of necessitating the upload of entire files onto our server. Additionally, the BUTLR format reduces the storage space of contact matrix files, not only through the binarization of files but also through the omission of redundant values. Perhaps the most striking example in this regard is that a 1-kb hg19 intrachromosomal contract matrix requires ~1 TB of storage for the tab-delimited matrix file and ~32GB for the coordinated list format. In comparison, the BUTLR format only requires ~11GB of storage, in addition to the benefit of random access.
To browse and visualize user-generated contact matrix files or any other datasets not hosted on our server, convert them to the BUTLR file format with BUTLRTools, a set of Perl scripts, available at Github. Ensure to download the entire folder instead of separate scripts as they are reliant on modules located in the same folder.
Table of Contents
I. Contact Matrix Formats: Tab-Delimited vs. Coordinated List
II. Output of Different Hi-C Pipelines (homerToMatrix.pl)
III. Convert to BUTLR (matrixToButlr.pl)
IV. Convert from BUTLR (butlrToMatrix.pl)
V. Upload to the 3D Genome Browser
VI. Troubleshooting Common Errors
I. Contact Matrix Formats: Tab-Delimited vs. Coordinated List
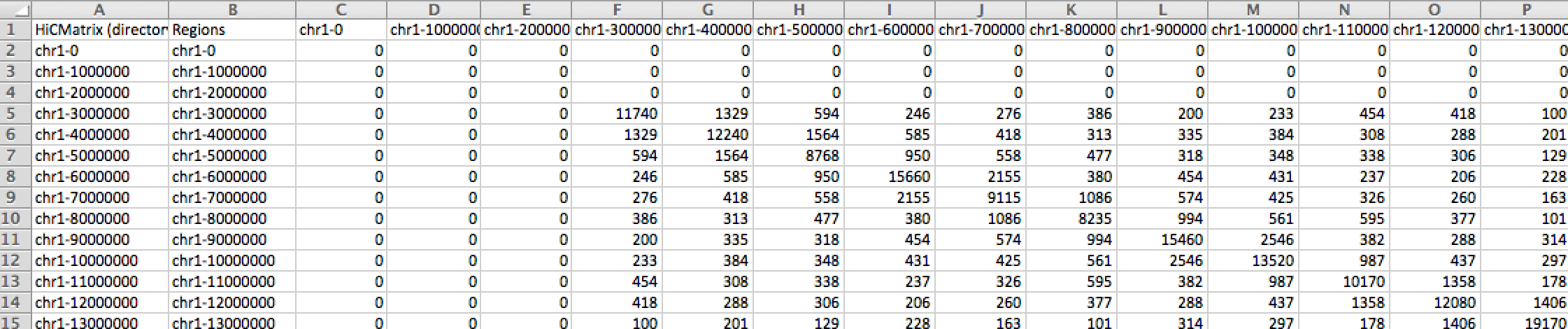
The tab-delimited text file encodes the entire contact matrix with each element in columns separated by tabs"\t" and each element in rows, newlines "\n". Hi-C datasets generated by the Bing Ren group, some of which are hosted under the 'DOWNLOAD' tab, are in the tab-delimited format. Below is a simplified example of a tab-delimited file format of a 5 × 5 contact matrix.24 7 3 0 0 7 16 10 5 0 3 10 19 12 4 0 5 12 21 9 0 0 4 9 17
This contact matrix stores intrachromosomal interactions. Notice that the values of the matrix are mirrored across its diagnonal, which makes sense biologically as the number of contacts between locus i and locus j will equal those between locus j and locus i. This redundacy would be a source to be resolved by the BUTLR file format to reduce storage. The current matrix is 91 bytes.
The coordinated list is a form of representing the sparse matrix, a matrix in which most of the elements are zero. The contact matrix of the mammalian genomes is sparse, as most of the interactions are highly localized. Since most of the elements in the sparse matrix are the same, these elements are often not stored. The coordinated list stores a list of tuples (row, column, value) describing all nonzero elements in the sparse matrix. In addition, since the entire matrix does not to be stored anymore, the diagonally mirrored values could be also neglected in storage, as the contact information stored in (row i, column j) also reflects the value of (row j, column i). The Hi-C datasets generated and shared by the Erez Lieberman Aiden group, in particular the Rao et al 2014 paper, are in this format. Below is an example of a coordinated list of the tab-delimited matrix above. Let us assume that the resolution is 40kb.
0 0 24 0 40000 7 0 80000 3 40000 40000 16 40000 80000 10 40000 120000 5 80000 80000 19 80000 120000 12 80000 160000 4 120000 120000 21 120000 160000 9 160000 160000 17
The size of this example is 207 bytes, compared to the tab-delimited text example of 91 bytes. Astute readers may notice that this example is not a sparse matrix, but rather a dense matrix. In a dense matrix, a coordinate list may not reduce storage as the storing row and column positions may offset the omitted zeros. As noted, the mammalian genome contact matrices, however, are sparse matrices which would benefit with the coordinate list format.
II. Output of Different Hi-C Pipelines (homerToMatrix.pl)
As intimated from the previous section, different laboratory groups utilize different pipelines that yield contact matrices in different format. The Ren group pipeline outputs contact matrices stored as tab-delimited text format in separate files as divided by chromosomes (chr1.matrix for chromosome 1, chr2.matrix for chromosome 2, chr3.matrix for chromosome 3, etc). The datasets from Dixon et al 2012 paper are hosted under the 'DOWNLOAD' tab are in this format. Also described above, the Lieberman Aiden group provided the Rao et al 2014 datasets in the coordinated list format, also separated by chromosomes. Meanwhile, another established Hi-C pipeline from HOMER yields yet a third format, one tab-delimited file that contains all chromosomes and all their intra- and inter-chromosomal interactions. Click the figure below for more information.
In an effort to standardize the formats, the script
perl homerToMatrix.pl -m <homer matrix> -g <genome size file> [-o <output file prefix>]
This command will yield files with
[output file prefix].[chrom1].[chrom2].matrix, where [chrom1] provides the bins by rows and [chrom2] provides the bins by columns. Since interchromosomal matrices [output file prefix].[chrom1].[chrom2].matrix and [output file prefix].[chrom2].[chrom1].matrix are tranpose matrices that provide redudant interaction entries, only one file, where the size of [chrom1] (reflected by number of rows) will be greater than [chrom2] will be created. Additionally the command will output a matrix list file, [output file prefix].list, which would serve as an input in the script to convert to BUTLR,
III. Convert to BUTLR (matrixToButlr.pl)
The Perl script, perl matrixToButlr.pl -g <genome size file> -m <matrix list file> -a <genome assembly> -r <resolution> [-h <row number where matrix begins (1-based)>][-o <output filename>]
Genome Size File
Two-column, tab-delimited file with column 1 containing chromosome names and column 2, size of the corresponding chromosome. These files are often available as[assembly].chrom.sizes through browsing the genome assembly datasets on the UCSC Genome Browser Downloads page (using Google is faster). If the file is not available, one could be created using the twoBitInfo program from the UCSC Utilities on the [assembly].2bit files (which itself could be converted from fasta files through chromInfo under each database named after genome assemblies. Below is an example of genome size file for hg19:
chr1 249250621 chr2 243199373 chr3 198022430 chr4 191154276 chr5 180915260 chr6 171115067 chr7 159138663 chrX 155270560 chr8 146364022 chr9 141213431 chr10 135534747 chr11 135006516 chr12 133851895 chr13 115169878 chr14 107349540 chr15 102531392 chr16 90354753 chr17 81195210 chr18 78077248 chr20 63025520 chrY 59373566 chr19 59128983 chr22 51304566 chr21 48129895
Matrix List File
Intrachromosomal Matrices Only: Two-column, tab-delimited file with column 1 containing chromosome names and column 2, filenames of each contact matrix (tab-delimited text format) for each corresponding chromosome. Example (matrix.list):
chr1 /directory-to-file/chr1.matrix chr2 /directory-to-file/chr2.matrix chr3 /directory-to-file/chr3.matrix chr4 /directory-to-file/chr4.matrix chr5 /directory-to-file/chr5.matrix chr6 /directory-to-file/chr6.matrix chr7 /directory-to-file/chr7.matrix chrX /directory-to-file/chrX.matrix chr8 /directory-to-file/chr8.matrix chr9 /directory-to-file/chr9.matrix chr10 /directory-to-file/chr10.matrix chr11 /directory-to-file/chr11.matrix chr12 /directory-to-file/chr12.matrix chr13 /directory-to-file/chr13.matrix chr14 /directory-to-file/chr14.matrix chr15 /directory-to-file/chr15.matrix chr16 /directory-to-file/chr16.matrix chr17 /directory-to-file/chr17.matrix chr18 /directory-to-file/chr18.matrix chr20 /directory-to-file/chr20.matrix chr19 /directory-to-file/chr19.matrix chr22 /directory-to-file/chr22.matrix chr21 /directory-to-file/chr21.matrix
Inclusion of Interchromosomal Matrices: Three-column, tab-delimited file with column 1 containing name of
chrom1, column 2, chrom2 and column 3, filenames of each contact matrix (tab-delimited text format) for interaction between chrom1 and chrom2. Since interchromosomal matrices [file prefix].[chrom1].[chrom2].matrix and [file prefix].[chrom2].[chrom1].matrix are tranpose matrices that provide redudant interaction entries, only one file out of both is needed. Currently, the script will only support interchromosomal matrix inputs where number of rows > number of columns, so its transpose matrix will cause an error. While the current 3D Genome Browser does not support the visualization of interchromosomal interactions, there are plans to develop this feature in the future. In addition, the current BUTLR supports the encoding of interchromosomal interaction matrices, which allows random access as well as binary and redundancy compression.
chr1 chr1 /directory-to-file/chr1.chr1.matrix chr1 chr2 /directory-to-file/chr1.chr2.matrix chr1 chr3 /directory-to-file/chr1.chr3.matrix chr1 chr4 /directory-to-file/chr1.chr4.matrix chr1 chr5 /directory-to-file/chr1.chr5.matrix chr1 chr6 /directory-to-file/chr1.chr6.matrix chr1 chr7 /directory-to-file/chr1.chr7.matrix chr1 chrX /directory-to-file/chr1.chrX.matrix chr1 chr8 /directory-to-file/chr1.chr8.matrix chr1 chr9 /directory-to-file/chr1.chr9.matrix chr1 chr10 /directory-to-file/chr1.chr10.matrix chr1 chr11 /directory-to-file/chr1.chr11.matrix chr1 chr12 /directory-to-file/chr1.chr12.matrix chr1 chr13 /directory-to-file/chr1.chr13.matrix chr1 chr14 /directory-to-file/chr1.chr14.matrix chr1 chr15 /directory-to-file/chr1.chr15.matrix chr1 chr16 /directory-to-file/chr1.chr16.matrix chr1 chr17 /directory-to-file/chr1.chr17.matrix chr1 chr18 /directory-to-file/chr1.chr18.matrix chr1 chr20 /directory-to-file/chr1.chr20.matrix chr1 chr19 /directory-to-file/chr1.chr19.matrix chr1 chr22 /directory-to-file/chr1.chr22.matrix chr1 chr21 /directory-to-file/chr1.chr21.matrix chr2 chr2 /directory-to-file/chr2.chr2.matrix chr2 chr3 /directory-to-file/chr2.chr3.matrix chr2 chr4 /directory-to-file/chr2.chr4.matrix chr2 chr5 /directory-to-file/chr2.chr5.matrix chr2 chr6 /directory-to-file/chr2.chr6.matrix chr2 chr7 /directory-to-file/chr2.chr7.matrix chr2 chrX /directory-to-file/chr2.chrX.matrix chr2 chr8 /directory-to-file/chr2.chr8.matrix chr2 chr9 /directory-to-file/chr2.chr9.matrix chr2 chr10 /directory-to-file/chr2.chr10.matrix chr2 chr11 /directory-to-file/chr2.chr11.matrix chr2 chr12 /directory-to-file/chr2.chr12.matrix chr2 chr13 /directory-to-file/chr2.chr13.matrix chr2 chr14 /directory-to-file/chr2.chr14.matrix chr2 chr15 /directory-to-file/chr2.chr15.matrix chr2 chr16 /directory-to-file/chr2.chr16.matrix chr2 chr17 /directory-to-file/chr2.chr17.matrix chr2 chr18 /directory-to-file/chr2.chr18.matrix chr2 chr20 /directory-to-file/chr2.chr20.matrix chr2 chr19 /directory-to-file/chr2.chr19.matrix chr2 chr22 /directory-to-file/chr2.chr22.matrix chr2 chr21 /directory-to-file/chr2.chr21.matrix chr3 chr3 /directory-to-file/chr3.chr3.matrix chr3 chr4 /directory-to-file/chr3.chr4.matrix chr3 chr5 /directory-to-file/chr3.chr5.matrix chr3 chr6 /directory-to-file/chr3.chr6.matrix chr3 chr7 /directory-to-file/chr3.chr7.matrix chr3 chrX /directory-to-file/chr3.chrX.matrix chr3 chr8 /directory-to-file/chr3.chr8.matrix chr3 chr9 /directory-to-file/chr3.chr9.matrix chr3 chr10 /directory-to-file/chr3.chr10.matrix chr3 chr11 /directory-to-file/chr3.chr11.matrix chr3 chr12 /directory-to-file/chr3.chr12.matrix chr3 chr13 /directory-to-file/chr3.chr13.matrix chr3 chr14 /directory-to-file/chr3.chr14.matrix chr3 chr15 /directory-to-file/chr3.chr15.matrix chr3 chr16 /directory-to-file/chr3.chr16.matrix chr3 chr17 /directory-to-file/chr3.chr17.matrix chr3 chr18 /directory-to-file/chr3.chr18.matrix chr3 chr20 /directory-to-file/chr3.chr20.matrix chr3 chr19 /directory-to-file/chr3.chr19.matrix chr3 chr22 /directory-to-file/chr3.chr22.matrix chr3 chr21 /directory-to-file/chr3.chr21.matrix . . . chr22 chr22 /directory-to-file/chr22.chr22.matrix chr22 chr21 /directory-to-file/chr22.chr21.matrix chr21 chr21 /directory-to-file/chr21.chr21.matrix
Genome Assembly
Genome assembly of the Hi-C dataset/contact matrices. This field is important to display the correct UCSC Genome Browser Session. If the assembly is user-generated, please refer to the guide to create assembly hubs on UCSC. After the hub is created, simply copy and paste the URL to the UCSC Genome Browser Session ID textbox.Resolution of Contact Matrix
The resolution of the contact matrix, which could be provided as base-pairs (40000), kbps (40kb) or even Mkps (1Mb).Option: -h <rrow number where matrix begins (1-based)>
Specify the number of lines of headers (1-based). For example, with one line of header, the matrix will start at row number 2 (-h 2). Default: 1 (no header).
There is currently no options for establishing the number of columns where matrix begins (existence of row names). The script will assume that any additional columns at beginning compared to the number of bins calculated by chromosome size and matrix resolution are row names and disregard accordingly.
Example
Below is an example of creating BUTLR files from hg19 GM12878 at 40kb:perl matrixToButlr.pl -g hg19.chrom.sizes -m matrix.list -a hg19 -r 40kb -o GM12878.40kb.btr
Sanity Checks
To minimize errors, ensure that the matrix is legal. Manually calculate the expected number of bins withMake sure that this number matches the number of rows in the contact matrix
wc -l <chrom.matrix>
awk -F "\t" -v OFS="\t" '{print NF}' <chrom.matrix> | uniq
IV. Convert from BUTLR (butlrToMatrix.pl)
TBA
V. Upload to the 3D Genome Browser
TBA
VI. Troubleshooting Common Errors
TBA
